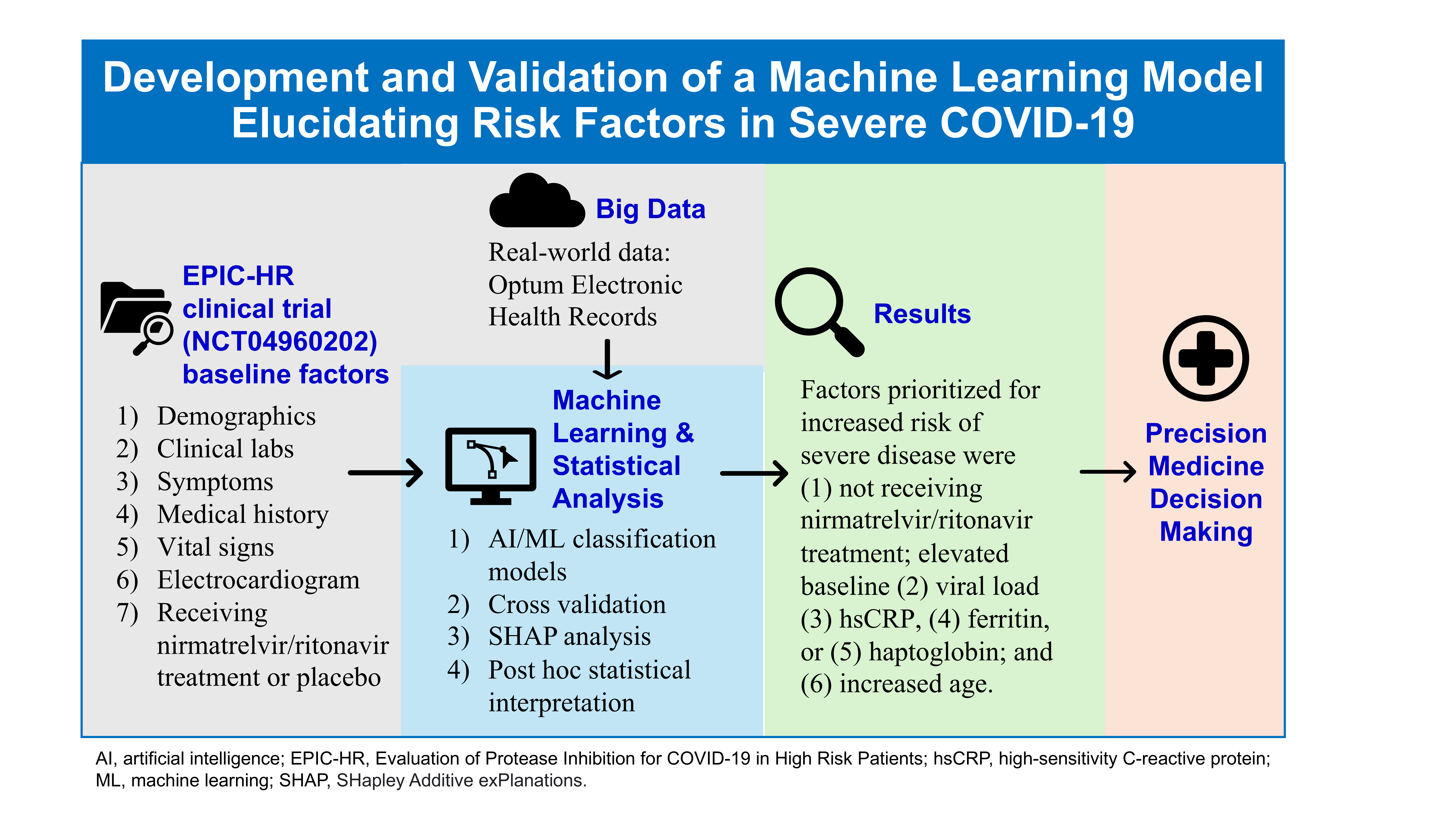
Volumes & Issues
Contact
For any inquiries regarding journal development, the peer review process, copyright matters, or other general questions, please contact the editorial office.
Editorial Office
E-Mail: biomedinfo@elspub.com
For production or technical issues, please contact the production team.
Production Team
E-Mail: production@elspub.com
About This Journal
Latest Articles
Editor's Choice
Top Downloaded
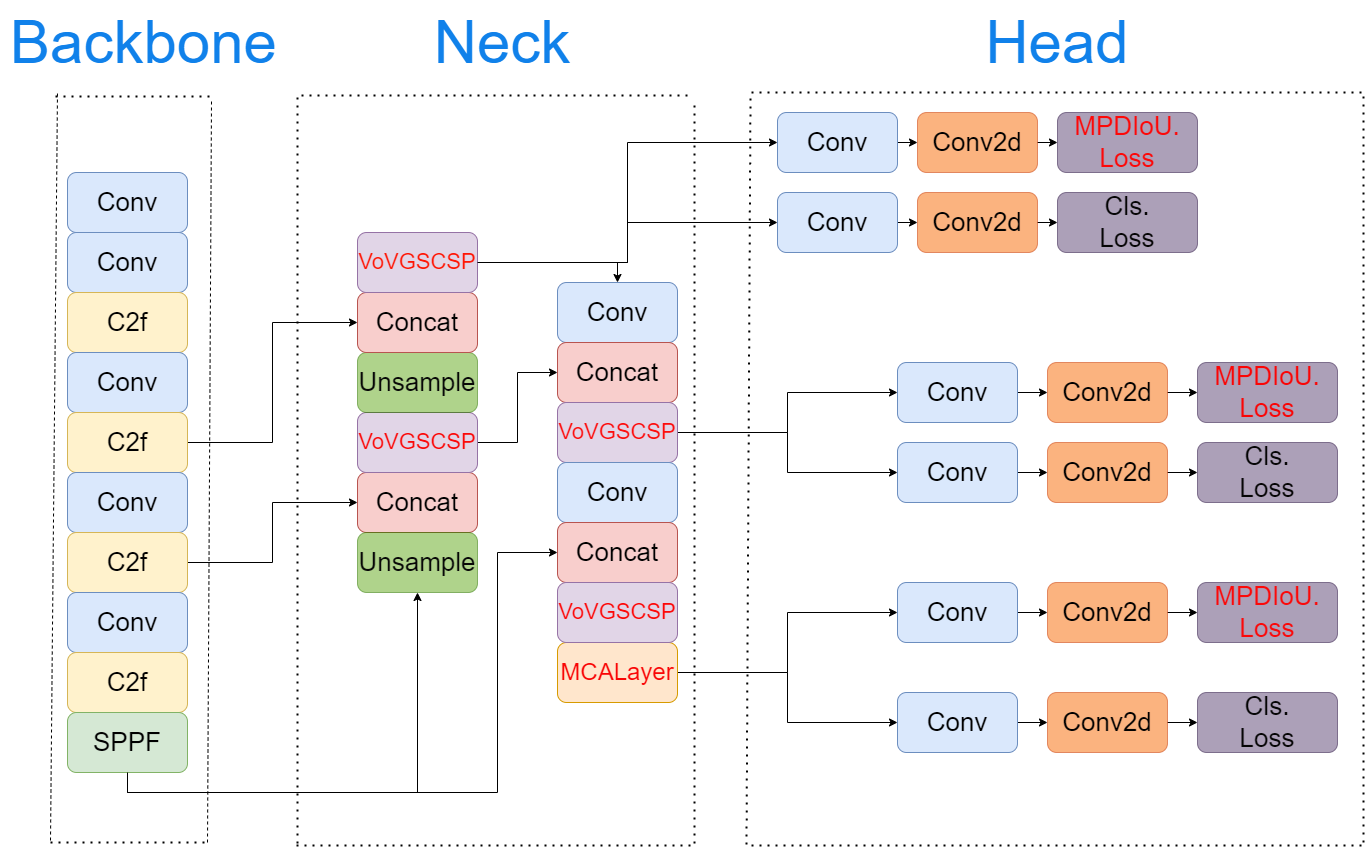
Micro-expression detection in ASD movies: a YOLOv8-SMART approach
DOI: 10.55092/bi20250002
Published: 25 Feb, 2025